Orthanc at UCLouvain
UCLouvain university (Belgium) stands proudly as the primary driving force behind the development and evolution of Orthanc. This versatile free and open-source ecosystem revolutionizes the way medical imaging data is managed, accessed, and utilized, empowering researchers, healthcare professionals, and scientific communities worldwide.

Research environment
Research on Orthanc is actively conducted within the Health Informatics Lab overseen by Sébastien Jodogne, the author of Orthanc. This laboratory is part of the Institute for Information and Communication Technologies, Electronics and Applied Mathematics (ICTEAM). The Computing Science and Engineering Department (INGI) of ICTEAM hosts and maintains the essential server infrastructure that is pivotal for the ongoing development and advancement of Orthanc, playing a crucial role in its evolution.
Main references
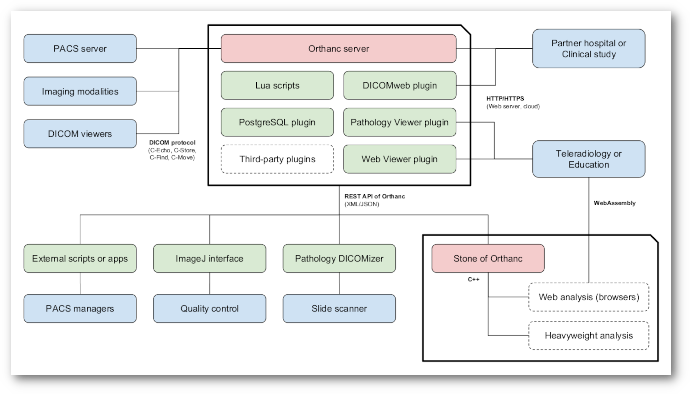
The Orthanc ecosystem for medical imaging
This paper reviews the components of Orthanc, a free and open-source, highly versatile ecosystem for medical imaging. At the core of the Orthanc ecosystem, the Orthanc server is a lightweight vendor neutral archive that provides PACS managers with a powerful environment to automate and optimize the imaging flows that are very specific to each hospital. The Orthanc server can be extended with plugins that provide solutions for teleradiology, digital pathology, or enterprise-ready databases.
Jodogne, Sébastien. Journal of Digital Imaging, Vol. 31, no.3, p. 341-352 (2018).
Demo server Read DocumentationOn the use of WebAssembly for rendering and segmenting medical images
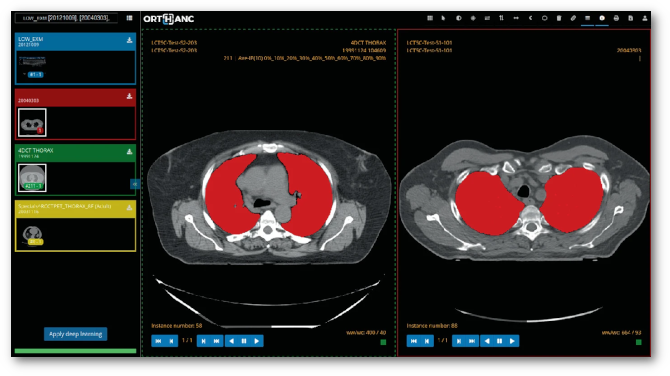
Rendering medical images is a critical step in a variety of medical applications, from diagnosis to therapy. In this paper, we show how the emerging WebAssembly standard can be used to share the same code base between heavyweight viewers and zero-footprint viewers. Moreover, we introduce a fully functional Web viewer that is entirely developed using WebAssembly and that can be used in research projects or in teleradiology applications. Finally, we demonstrate that deep convolutional neural networks for image segmentation can be executed entirely inside a Web browser thanks to WebAssembly.
Jodogne, Sébastien. Biomedical Engineering Systems and Technologies, Vol. 1814, p. 393-414 (2023).
Demo Stone Web viewer Read DocumentationOpen implementation of DICOM for whole-slide microscopic imaging
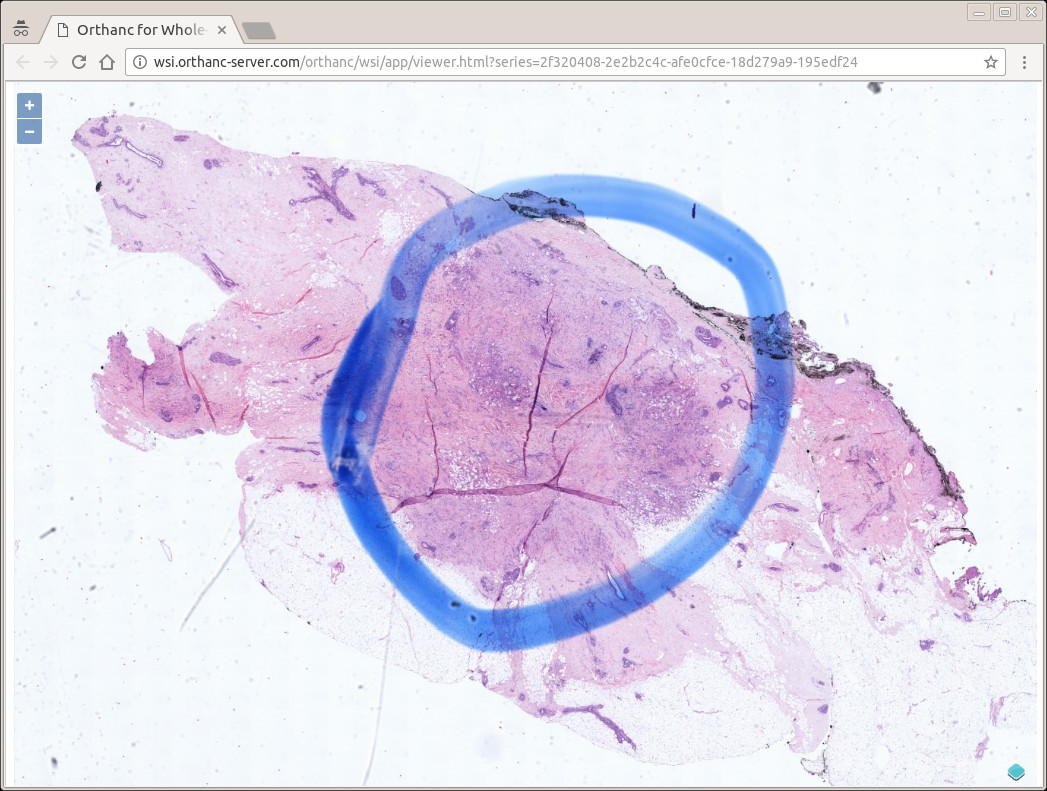
This paper introduces an open implementation of DICOM for whole-slide microscopic imaging, following Supplement 145 of the DICOM standard. The software is divided into two parts: (a) a command-line tool to convert an whole-slide image to the DICOM format, and (b) a zero-footprint Web interface to display such DICOM images. The software architecture leverages the DICOM server Orthanc. The entire framework is available as free and open-source software. The existence of this software supports the development of digital pathology and telepathology in clinical environments, featuring a smooth integration with existing EHR and PACS solutions.
Jodogne, Sébastien et al. Proc. of the 12th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications, p. 81-87 (2017).
Demo Read DocumentationMore publications
Combining Languages to Set a PACS on FHIR
Jodogne, Sébastien. Communications in Computer and Information Science, Vol. 2546, p. 314-334 (2025).
Read Documentation
Integrated and Interoperable Platform for Detecting Masses on Mammograms
Chatzopoulos, Edouard et al. Proc. of the 34th Medical Informatics Europe Conference (MIE 2024), Studies in Health Technology and Informatics, Vol. 316, p. 1103-1107 (2024).
Read Sources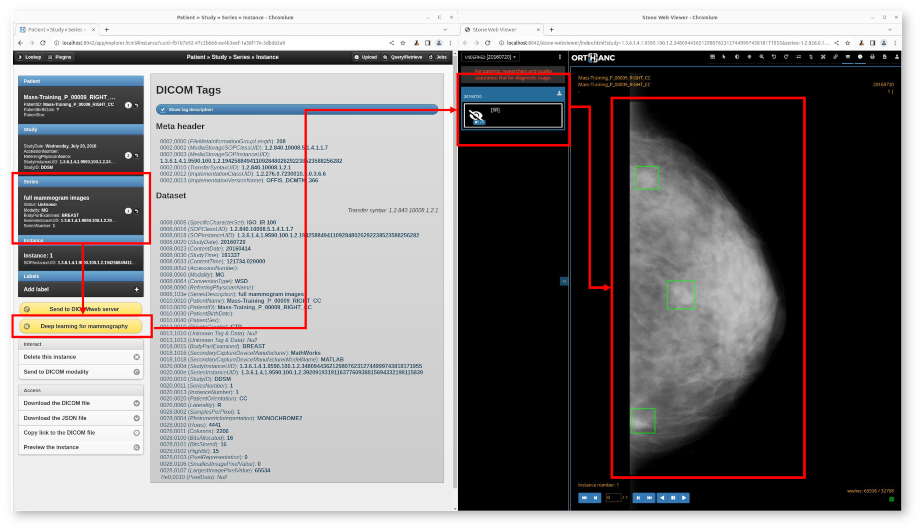
PARROT - A Versatile Platform for AI-Driven Image Segmentation and Dose Prediction
Sadre, Wei et al. Proc. of the 5th International Conference on Medical Imaging and Computer-Aided Diagnosis (MICAD 2024).
Read Homepage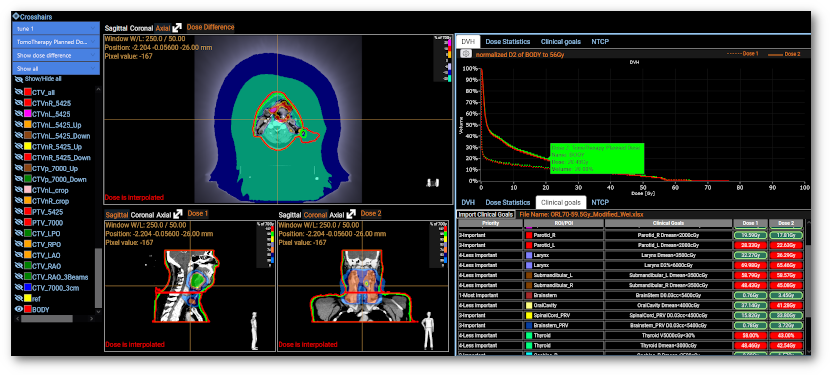
Setting a PACS on FHIR
Jodogne, Sébastien. Proc. of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC HEALTHINF), Vol. 2, p. 123-131 (2024).
Read Documentation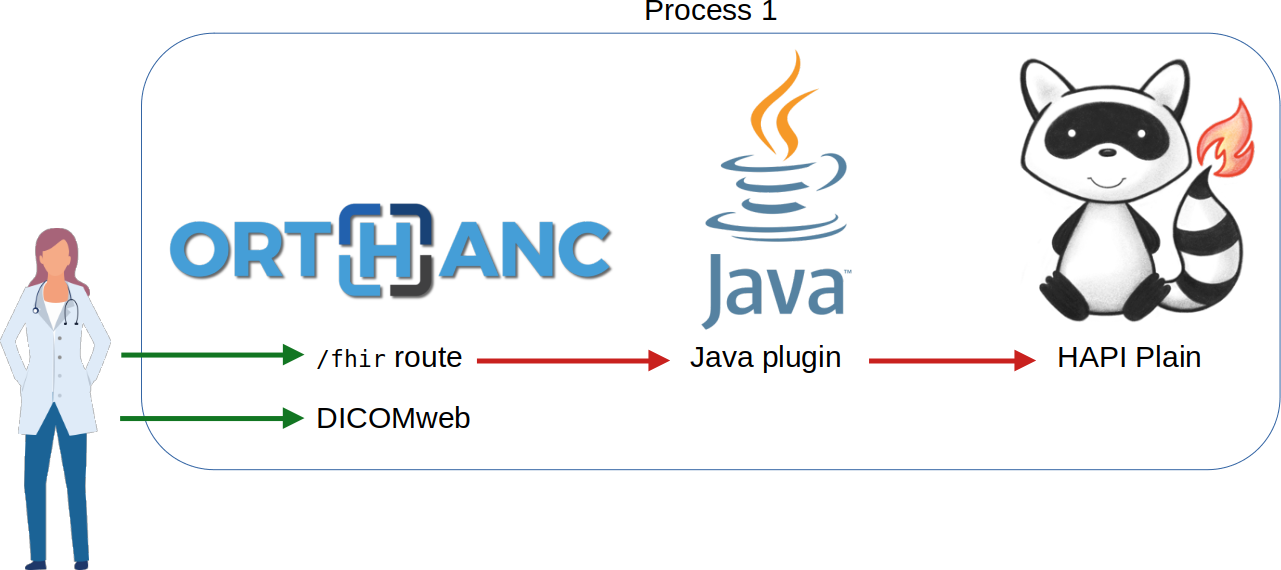
Serveur multimédia DICOM pour le partage des numérisations des collections anthropologiques
Semal, Patrick et al. 35e édition du Colloque du Groupement des Anthropologistes de Langue Française (GALF 2024).
Read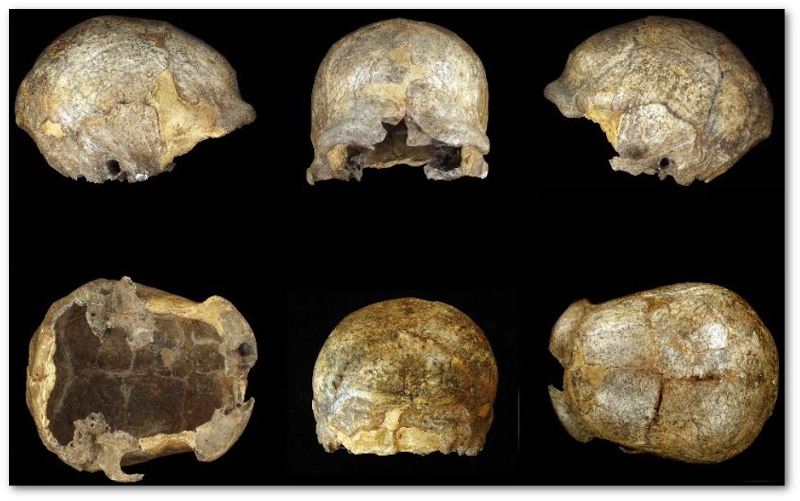
Open platform for the de-identification of burned-in texts in medical images using deep learning
Langlois, Quentin et al. Proc. of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC BIOIMAGING), Vol. 1, p. 297-304 (2024).
Read Sources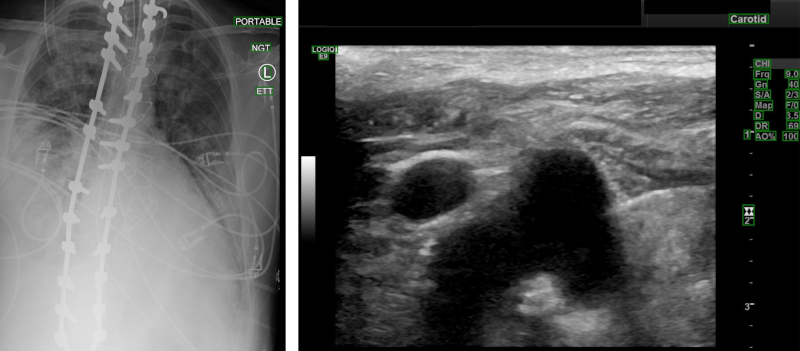
Interoperable encoding and 3D printing of anatomical structures
Fierens, Amaury et al. ICBRA '23: Proceedings of the 10th International Conference on Bioinformatics Research and Applications, p. 20-26 (2023).
Read Documentation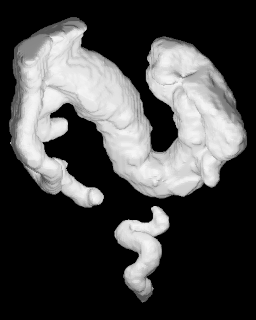
Client-side application of deep learning models through teleradiology
Jodogne, Sébastien. Medical Informatics Europe, Studies in Health Technology and Informatics, Vol. 302, no.1, p. 997-1001 (2023).
Read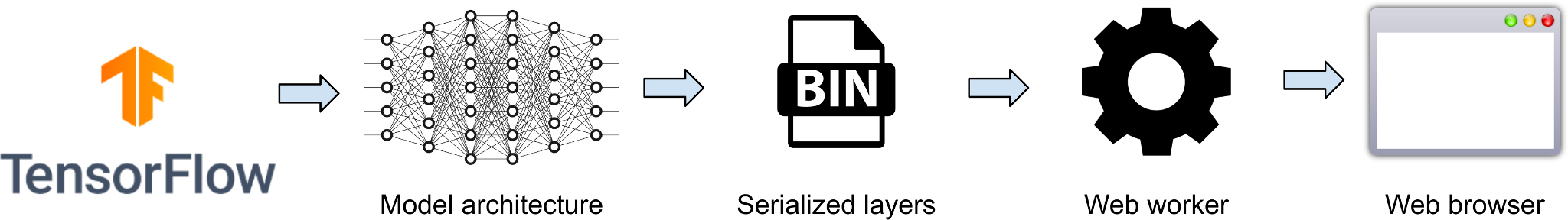
Simple, effective deployment of Web viewers for medical imaging in an open platform
Jodogne, Sébastien. EuSoMII Annual Meeting, AI: Connecting the dots (2023).
Abstract OHIF documentation VolView documentation
On the use of DICOM as a storage layer for IIIF
Jodogne, Sébastien. DaSCHCon on IIIF, 3D & Interoperability (2023).
Abstract Documentation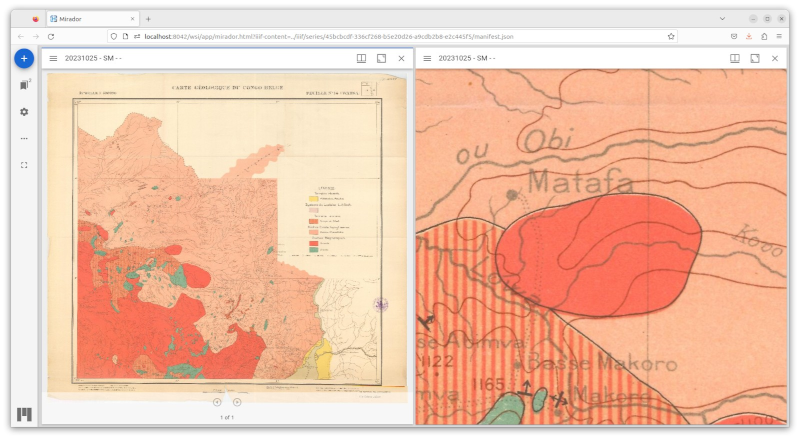
Rendering medical images using WebAssembly
Sébastien Jodogne. Proc. of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC BIOIMAGING), Vol. 2, p. 43-51 (2022).
Read Documentation
Étude de faisabilité : l'utilisation de l'imagerie médicale en éducation thérapeutique en radiothérapie
Kirkove, Delphine et al. Cancer Radiothérapie, Vol. 26, no. 8, p. 1034-1044 (2022).
Read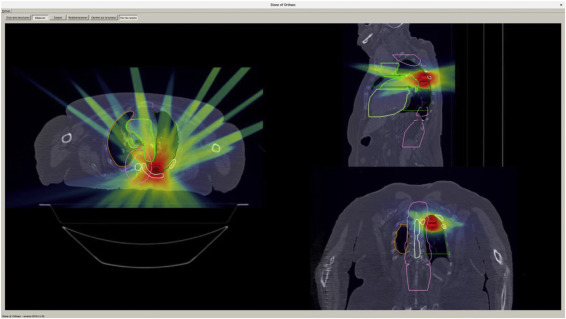
Federated learning for heart segmentation
Misonne, Thibaud and Jodogne, Sébastien. IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP) (2022).
Read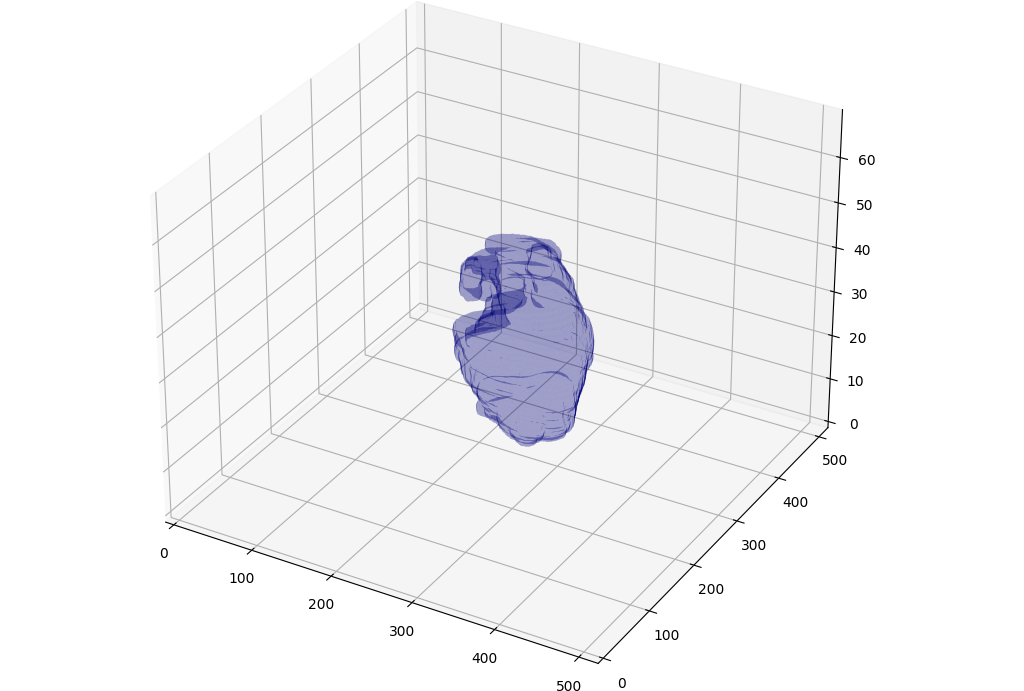
Automatically publishing medical images from a filesystem as a DICOM server
Insights into Imaging, Vol. 13, no. S2, p. 7 (2021).
Read Documentation
Importing and serving open-data medical images to support artificial intelligence research
Insights into Imaging, Vol. 13, no. S2, p. 6 (2021).
Read Documentation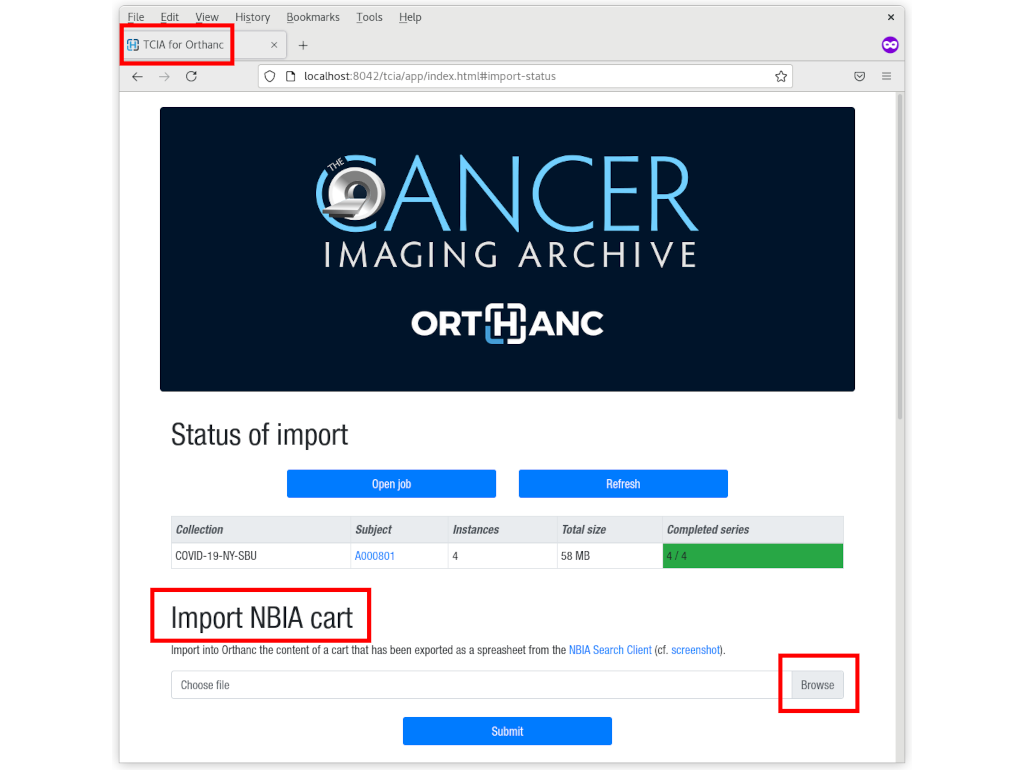
Automating DICOM retrieve from PACS with free and open-source software
Annual Congress of the European Association of Nuclear Medicine, European journal of nuclear medicine and molecular imaging, Vol. 45, no. S1, p. 298 (2018).
ReadAutomating de-identification and sharing DICOMs with a free and open-source software
Annual Congress of the European Association of Nuclear Medicine, European journal of nuclear medicine and molecular imaging, Vol. 45, no. S1, p. 299 (2018).
ReadOrthanc: A lightweight, RESTful DICOM server for healthcare and medical research
Jodogne, Sébastien et al. Proc. of the IEEE 10th International Symposium on Biomedical Imaging, Vol. 1, no.1, p. 190-193 (2013).
Read Documentation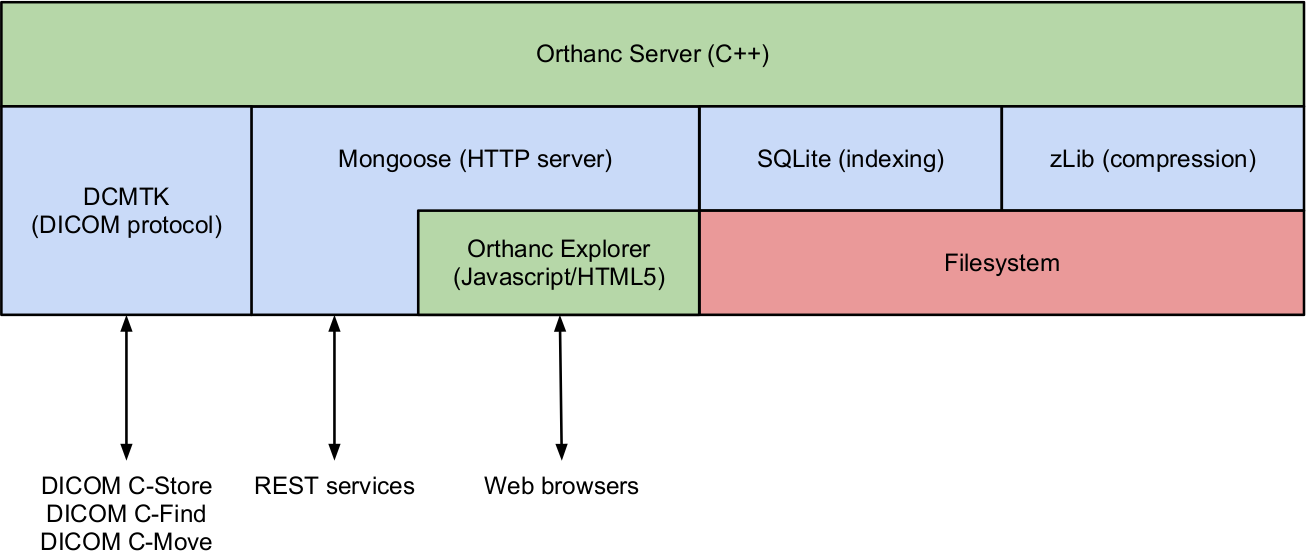
© 2021-2024, Sébastien Jodogne, ICTEAM, UCLouvain